Export Sub-Tomograms
This dialog allows you to reconstruct “sub-tomograms”, i. e. small, cubic volumes centered around a set of 3D coordinates you provide. The pixel size of these reconstructions is usually much smaller than the one used for full tomogram reconstruction because the volumes are later aligned on a common target and averaged to obtain better signal. Warp also reconstructs a 3D CTF volume for each particle to be used in RELION’s refinement procedure.
If you’re only starting to incorporate Warp into your tomography pipeline, a few things can go wrong at this step. The first sanity check to do after sub-tomogram export is to average all the volumes and hopefully observe a white blob in the middle of the average. This way you know that the particle positions coincide with the actual particles, as opposed to being off if something ist wrong. The easiest way to get the average is to use RELION’s relion_reconstruct (or relion_reconstruct_mpi for the parallelized version). The command looks something like this:
relion_reconstruct --i particles_from_warp.star --o rec.mrc --3d_rot --ctf
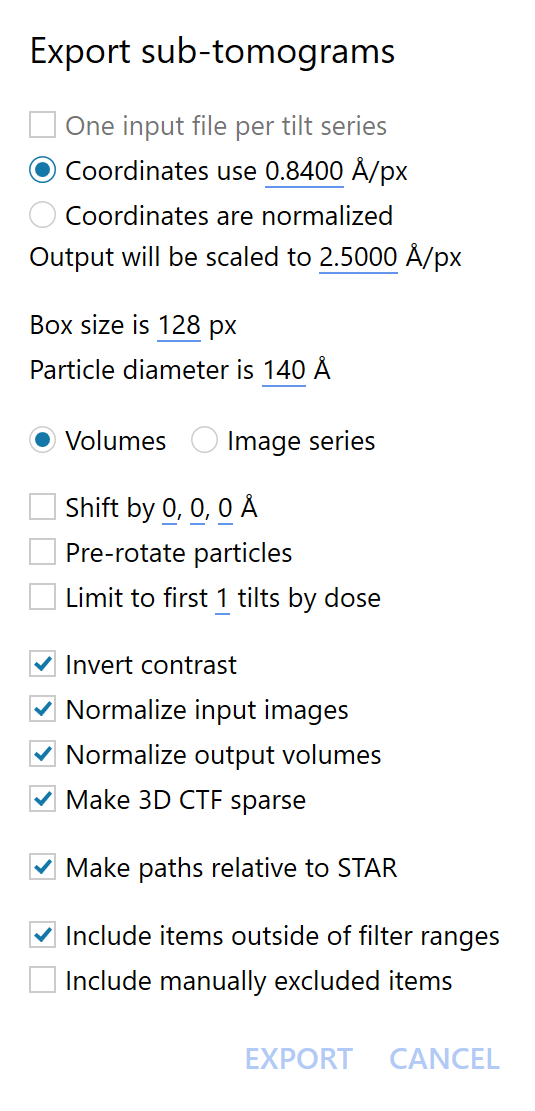
Before you see the actual dialog, Warp will ask you to provide a file with particle coordinates. Only RELION’s STAR format is currently supported. The file must contain at least the rlnCoordinateX/Y/Z columns. If rlnOriginX/Y/Z are specified, Warp will consider them, and the values will be reset to 0 in the STAR file Warp generates. You can use either a single file for all tilt series or, if you have one file per tilt series (e. g. after template matching) with names formatted as [series name]_suffix.star, you can point Warp to one of those files and it will figure out the suffix automatically and find all remaining files in the same folder. In the second case, One input file per tilt series will become checked. If you’d like to use just the file you selected and reconstruct sub-tomograms from only one tilt series, you can uncheck it.
Coordinates use…: If the coordinates come from RELION, they are in pixels. Because you might want to switch the pixel size occasionally, Warp handles all coordinates in Angstrom internally. Thus, you need to specify the pixel size used in that particular STAR file. It is assumed to be the same for rlnCoordinateX/Y/Z and rlnOriginX/Y/Z.
Coordinates are normalized: If the coordinates come from Warp’s template matching, they are normalized to a range of [0; 1]. Warp will automatically scale them back to the physical tomogram dimensions if you check this option.
Output will be scaled to…: The target pixel size to which the tilt series data will be scaled prior to reconstruction. It can’t be smaller than the size dictated by the Bin parameter in the Input settings section. The maximum resolution achievable with the data will be twice the pixel size.
Box size is…: The sub-tomogram box size in pixels. Warp avoids CTF aliasing by calculating the necessary minimum box size for the particle’s defocus value and the expected maximum resolution, as dictated by the pixel size, and then crops the volume back to the value you specified. For high-defocus data and small boxes, this can make the high-resolution Thon rings invisible. Don’t worry, this is fine. In fact, it is much finer than previous methods that would have made the data useless beyond the aliasing-free resolution.
Particle diameter is…: The particle diameter should be roughly the same you will use later in RELION, otherwise it might complain about incorrect normalization.
Volumes / Image series: In addition to reconstructing 3D sub-tomograms, Warp can also generate a series of 2D tilt images for each particle. There is currently no use for this data type in RELION, unless you have giant particles with a lot of signal and would like to refine the particle tilt images completely independently like in EMAN 2.3. This is probably not a good idea.
Shift by…: You can shift all particle positions by the same amount in 3D. If the input STAR file contains refined particle orientations (rlnAngleRot/Tilt/Psi columns), the shift will be made in the refined map’s reference frame. Use this if you have aligned all particles on a common part of the protein, and want to e. g. shift the map center to a ligand to analyze it further. Note that the shift values are in Angstrom.
Pre-rotate particles: If refined particle orientations (rlnAngleRot/Tilt/Psi columns) are available, the reconstructions can be pre-rotated to their common reference frame. If you were to average the resulting sub-tomograms without considering any rotations (e. g. simply sum them up in Matlab), you would get the refined map. This can be useful if you’re developing a new method to analyze the particles (e. g. variance, 3D PCA etc.) and don’t want to bother with particle orientations. The 3D CTFs will be pre-rotated to the same reference frame, too.
Limit to first N tilts by dose: The reconstructions or particle tilt series will contain only data from the first N tilts, sorted by dose. This can be useful if you think the late tilts don’t contain any information that might help your analysis.
Invert contrast: Multiply the input data by -1 to invert the contrast. Following standard conventions, black particles from cryo data will become white if this is enabled.
Normalize input images: The scaled tilt images will be normalized to mean = 0, standard deviation = 1 prior to reconstruction.
Normalize output volumes: Normalizes the reconstructions so that the volume outside the specified particle diameter is mean = 0, standard deviation = 1. This is required for RELION.
Make 3D CTF sparse: Don’t use this just yet! It requires a patch for RELION that isn’t public yet. If enabled, every voxel of the 3D CTF with a value below 0.01 will be explicitly set to 0. This allows the patched RELION algorithms to completely exclude them from calculations, saving a lot of time.
Make paths relative to STAR: Paths to the reconstructed volumes and 3D CTFs will be made relative to wherever you choose to save the output STAR file. Usually a good idea.
After clicking Export, Warp will ask you to specify the name and location of the output STAR file that will contain the paths of all volumes and their metadata. Use this file as input for RELION.